The development of effective treatments for chronic diseases, such as heart disease, diabetes, cancer, and Alzheimer’s, is a key global healthcare challenge. These conditions are primary drivers of healthcare spending and have a deleterious effect on quality of life and mortality. Fortunately, they are inextricably linked to modifiable health-related behavioral risk factors such as smoking, poor diet, and lack of physical activity. Mobile health technology [8] provides a novel tool for supporting lifestyle modifications via behavioral interventions that can provide personalized guidance to patients, thereby reducing their risk factors and improving health outcomes.
While app-based therapies based on self-report data (i.e. EMA) are increasingly available, passive sensing from wearables holds the promise of continuously monitoring key physiological and contextual variables, thereby providing long-term insight into patterns of behavior and risk at low burden to participants. We are developing novel biomarkers from wearable sensors to capture physiological and contextual variables, and machine learning methods for modeling longitudinal data. We work with the mDOT and MD2K [3] centers and have a broad set of collaborators in engineering, behavioral science, and statistics, with a focus on smoking, sedentary behavior, and substance abuse. We currently work in three research areas:
- Modeling and imputing missing measurements in Mobile Health data
- Biomarkers for physiological and contextual risk factors
- Predictions of adverse outcomes from event data
Modeling and imputing missing measurements in Mobile Health data
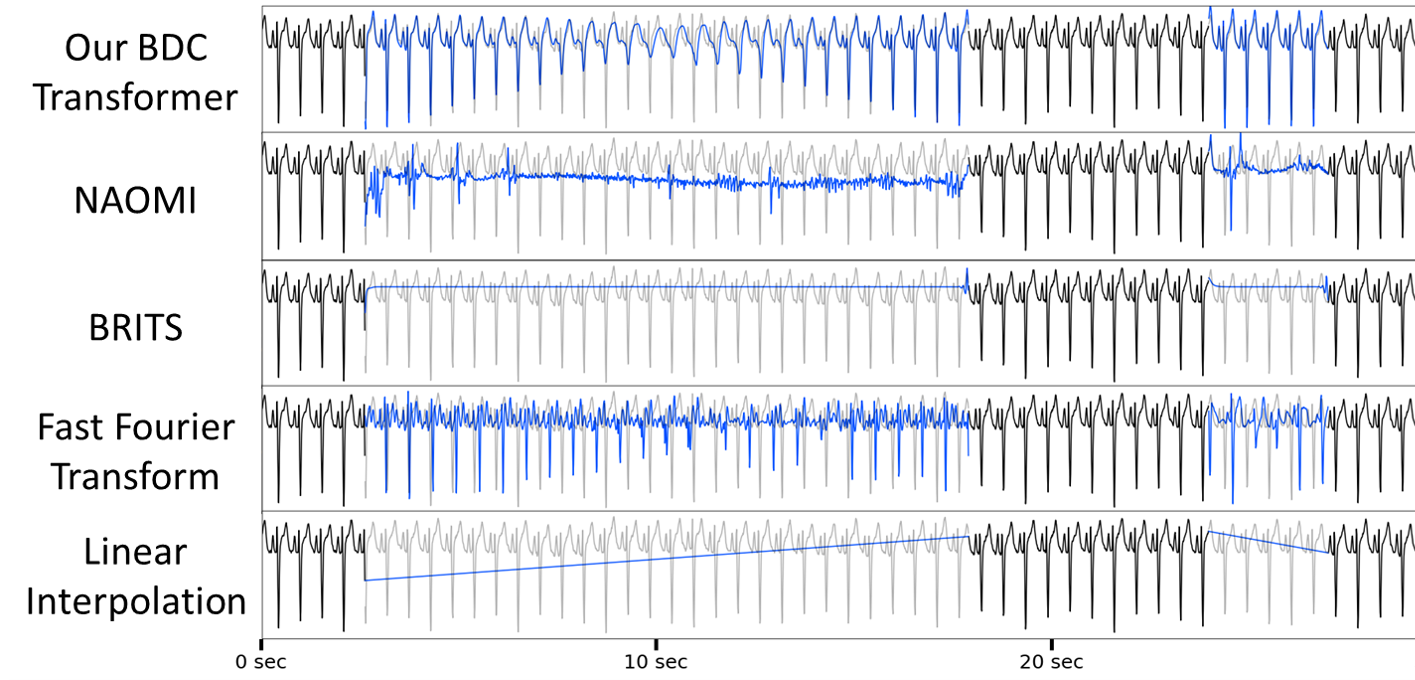
Problems with missing data are endemic to clinical and mobile health applications. We are developing principled strategies for imputing missing observations from temporal data in the context of the hierarchical computations that naturally arise in mHealth biomarker inference. We have recently addressed the problem of modeling the mean function of a counting process in mobile survey data with missing counts [6], and proposed novel mHealth pulsative signal (e.g. ECG and PPG) imputation challenge, PulseImpute. Prior work time-series imputation work fails in our setting, but our BDC Transformer demonstrates the potential of exploiting signal structure for pulsative signal imputation. Pulsative signal imputation over long gaps as well as reconstruction of features tied to Diagnosis are key unsolved mHealth challenges, and we hope that our challenge can drive future work in this critical and unique challenge.
Biomarkers for physiological and contextual risk factors
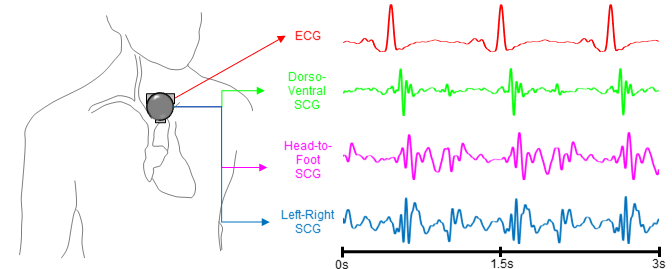
We are collaborating with the Inan Lab in developing novel biomarkers for hemodynamic parameters, such as pulmonary capillary wedge pressure, and in developing predictive models for heart failure (HF) clinical status (i.e. compensated vs decompensated). Decompensation in HF is a key driver of rehospitalization, resulting in increased mortality risk and cost of care. Using an implantable device to measure pulmonary artery pressure and titrate treatment has been shown to reduce the risk of decompensation, but the procedure is invasive, expensive, and unsuitable for some patients. We are exploring ballistocardiography and seismocardiography as a means to estimate heart failure status by measuring and analyzing the small vibrations that propagate through the body due to the mechanical action of the heart. Ballistocardiography (BCG) uses a modified weighing scale, and we have demonstrated classification of HF decompensation at an AUC of 0.78 using BCG data collected in a home environment [1]. Seismocardiography uses an accelerometer placed on the chest and is currently under investigation.
Predictions of adverse outcomes from event data
Many common temporal data sources in clinical and mobile health applications consist of event data, in which measurements occur at arbitrary, irregularly-distributed time points. Visits to the clinic for assessment, which are common in treating conditions such as glaucoma or autism, rarely occur on a fixed schedule. Likewise, in mobile health applications it is common to perform Ecological Momentary Assessments (EMA) at randomly-chosen times throughout the day. We have developed two approaches to modeling such data:
- Latent state models based on Continuous Time HMM (CT-HMM)
- Deep learning models based on transformers
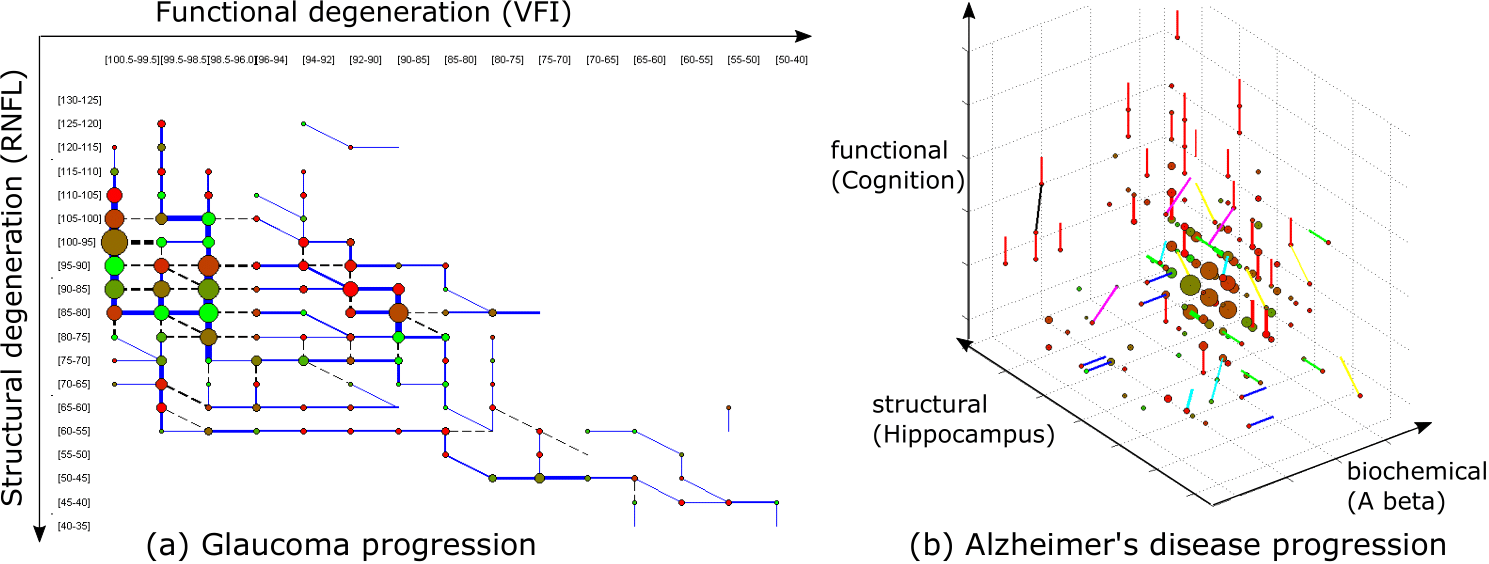
A CT-HMM is an HMM in which both the transitions between hidden states and the arrival of observations can occur at arbitrary (continuous) times, rather than on a fixed clock. We have developed efficient parameter learning methods for CT-HMMs [4] and have demonstrated their utility in modeling the progression of glaucoma and Alzheimer’s disease (illustrated below). We have developed a spatiotemporal CT-HMM approach to modeling glaucoma progression, which can describe the temporal history of 2D visual field and retinal thickness maps [7] and accurately predict future patient status as a tool for treatment management. We have developed a predictive model for relapse in substance abuse by combining a CT-HMM model of latent behavioral states with a survival model for risk prediction [5].
Personnel
Georgia Tech
Project Funding
- NIH Awards: P41-EB028242-01A1, 7-R01-MD010362-03
- NSF Awards: NSF GRFP DGE-2039655, RI 1320348, BCS 1524565, RAISE 1936970
- Gifts from Xerox PARC and Intel Research
References
- V. B. Aydemir , S. Nagesh, M. M. H. Shandhi, J. Fan, L. Klein, M. Etemadi, J. A. Heller, O. T. Inan, and J. M. Rehg, Classification of Decompensated Heart Failure From Clinical and Home Ballistocardiography. IEEE Transactions on Biomedical Engineering, vol. 67, no. 5, pp. 1303–1313, 2020.
- Chatterjee, S. , Moreno, A., Lizotte, S. L., Akther, S., Ertin, E., Fagundes, C. P., ... & Kumar, S. (2020). SmokingOpp: Detecting the Smoking Opportunity Context Using Mobile Sensors. Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies, 4(1), 1-26.
- Kumar, S. , Abowd, G. D., Abraham, W. T., Al’Absi, M., Gayle Beck, J., Chau, D. H., Condie, T., Conroy, D.E., Ertin, E., Estrin, D., Ganesan, D., Lam, C., Marlin, B., Marsh, C.B., Murphy, S.A., Nahum-Shani, I., Patrick, K., Rehg, J.M., Sharmin, M., Shetty, V., Sim, I., Spring, B., Srivastava, M., Wetter, D. W. (2015). Center of excellence for mobile sensor data-to-knowledge (MD2K). Journal of the American Medical Informatics Association, 22(6), 1137–1142.
- Liu, Y. Y. , Li, S., Li, F., Song, L., & Rehg, J. M. (2015). Efficient learning of continuous-time hidden markov models for disease progression. In Advances in neural information processing systems (pp. 3600-3608).
- Dempsey, W. H. Dempsey, W. H., Moreno, A., Scott, C. K., Dennis, M. L., Gustafson, D. H., Murphy, S. A., & Rehg, J. M. (2017). iSurvive: an interpretable, event-time prediction model for mHealth. Proceedings of machine learning research, 70, 970.
- Moreno, A. , Wu, Z., Yap, J. R., Lam, C., Wetter, D., Nahum-Shani, I., ... & Rehg, J. M. (2020). A Robust Functional EM Algorithm for Incomplete Panel Count Data. Advances in Neural Information Processing Systems, 33.
- Nagesh, S., Moreno, A., Ishikawa, H., Wollstein, G., Schuman, J. S. (2019). A Spatiotemporal Approach to Predicting Glaucoma Progression Using a CT-HMM. Machine Learning for Healthcare Conference (pp. 140-159).
- Rehg, J. M. , Murphy, S. A., & Kumar, S. (2017). Mobile Health: Sensors, Analytic Methods, and Applications. Springer.


